

 |
 |

NOTE: The error message "Operation failed!" means a suspicious command. Please send a message to administrator and explain the problem.
ExAtlas has been developed in the Laboratory of Genetics and Genomics at the
National Institute of Aging (NIA/NIH).
How to cite ExAtlas: Sharov, A.A., Schlessinger, D., and Ko, M.S.H. 2015. Exatlas: An
interactive online tool for meta-analysis of gene expression data.
J. Bioinform. Comput. Biol., DOI: 10.1142/S0219720015500195.
The workflow in ExAtlas is shown in Fig.1 below.
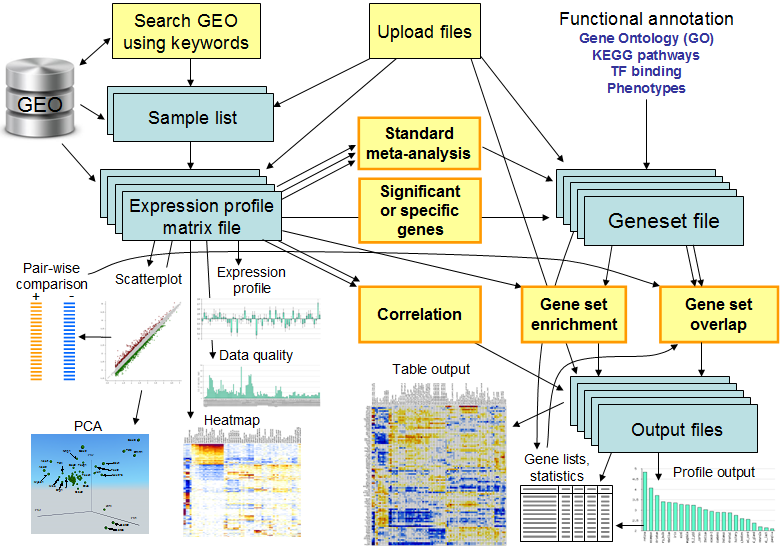
Fig. 1. Workflow in ExAtlas - software for gene expression meta-analysis.
For example, a user may search GEO database for specific terms such as "kidney", "muscle", or "T-cells", and the software provides information on samples where these terms are found. The user then selects samples from the list and the software generates a gene expression profile matrix. ExAtlas can evaluate the quality of data and then low-quality samples can be removed. Alternatively, expression profile data can be uploaded manually. The gene expression profile matrix can then be used for ANOVA, pair-wise comparison between tissues or cell types, Principal Component Analysis (PCA), making scatter-plots, expression profiles of individual genes, and heatmaps. Several gene expression matrices (e.g., GNF data on tissue/organ expression profiles) are pre-loaded in the software as public resources and are available to every user. Each gene expression matrix can be compared using correlation analysis with any other expression matrix.
Another important type of data is a gene set (or "geneset"). Each geneset file combines multiple genesets. The ExAtlas software stores many preloaded public geneset files, including Gene Ontology (GO), KEGG pathways, and BIOCARTA pathways. Gene set enrichment analysis (PAGE) is used to compare a gene expression matrix file with a geneset file. It evaluates if genes that are upregulated or downregulated in each tissue or cell type are enriched in specific genesets (e.g., GO or KEGG). Another option is to generate a new geneset file that contains genes that are significantly upregulated or downregulated in each tissue or cell type. This geneset file can be then tested for geneset overlap. The overlap is evaluated using hypergeometric distribution. Results of analysis are presented as color-coded tables or bar chart profiles.
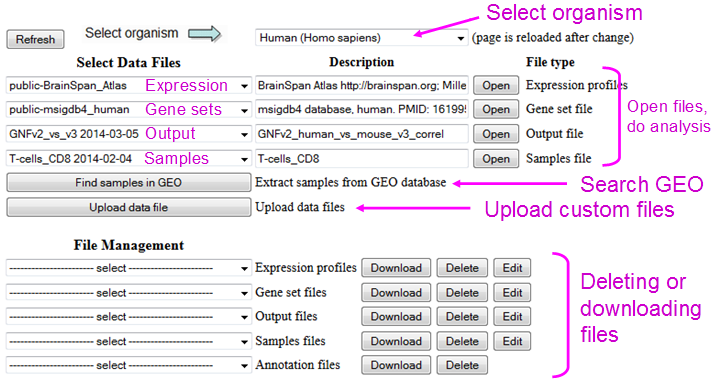
Fig. 2. The main menu in ExAtlas.
The main menu of the program (Fig. 2) includes pull-down menus for selecting data files (expression profile matrices, genesets, samples, and outputs), buttons for opening these files (used for visualization and for staring analysis), buttons to search GEO database and upload custom data files. The bottom portion of the main menu is used for downloading or deleting data files, as well as editing file names, file descriptions, column headers, and geneset names and descriptions. File editing also includes options to delete or copy selected expression profiles or genesets.
2. Register with ExAtlas
Registration is optional; you can login as guest (click the "Start using ExAtlas" button)
and do the analysis. However, registration has many benefits because you can keep your
profile, uploaded files, and save results of analysis for future sessions. We will not use
your e-mail address except to notify you of a new feature of ExAtlas (less than 1 message per year)
and will not release it to any third party. To register
click on the "Register here" link on the front page.
3. How to use ExAtlas? Step-by-step instructions
List of tasks you can do with ExAtlas
When you saved the samples, these samples will be appear in the web page. Alternatively, you can select any previously save file with samples and open it from the main menu (Fig. 2). The next step is to edit the list of samples. Some samples can be deleted or copied to another file. An important step is to edit descriptions of samples, because samples with identical descriptions within the same data set (GSE series) are considered as replications and will be placed together. Thus, replication numbers or array ID numbers should be removed from sample descriptions. If a description is not clear, click on sample ID and check detailed description in the GEO database, then edit sample description in the list of samples. After the list is finalized, generate a combined matrix of all samples by clicking the button "Generate matrix". Although samples from different data series (GSE accession numbers) can be combined in one list of samples, in many cases it is better to save each data series separately, upload corresponding data from GEO, and later combine data series using batch-normalization method (see Edit files). Downloading and processing the data takes some time. Thus, the "interruption screen" appears (see Fig. 3):
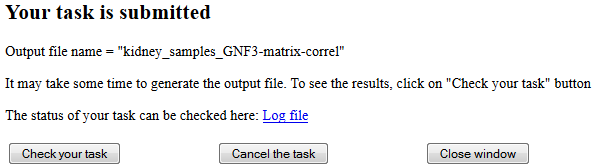
Fig. 3. Interruption screen is used for long computational tasks.
In this window you can check your task, cancel the task, or close the window without cancelling the task. The link to "Log file" is provided so that you can check the status of your task. Keep reloading the log file to see changes. If you click "Check your task" but it is not finished, then the screen will say "Your task is not finished!". Results will be shown when the task is finished. If data comes from different array platforms, expression profiles are combined based on gene symbol, and if multiple probes are available for a gene, then the best probe is used with either higher statistical significance (F-statistics) or higher average signal intensity (if there are no replications). However, if all samples are obtained with the same array platform, then redundant probes are not removed; and thus, a gene can be represented by multiple probes.
If you cannot find a specific data set, which you know exists in GEO, this may have resulted from data filtering. Your data set may have been filtered out because the array platform type is a cDNA array, tiling array, genomic array, exon array, non-matching species, or RNA-seq. If you believe that the data was filtered out by mistake, please send a note to webmaster. Currently, the GEO database has no uniform format for processed RNA-seq data, and thus, automated download is not possible. However, you can upload RNA-seq data (e.g., from Cufflinks) manually using "Upload data file" button (Fig. 2).
3.2. Open gene expression data and statistical results (ANOVA)
If you select a file with gene expression profiles in the main menu (Fig. 2) and then click the
button "Open", a new screen (or tab) will open in your browser, which allows you to display
expression profiles in various ways (Fig. 4). If you open this file for the first time after uploading,
then you may need to wait till the statistical analysis is finished. If the data contains too
many columns, the interruption screen (Fig. 3) may appear while the analysis is performed.
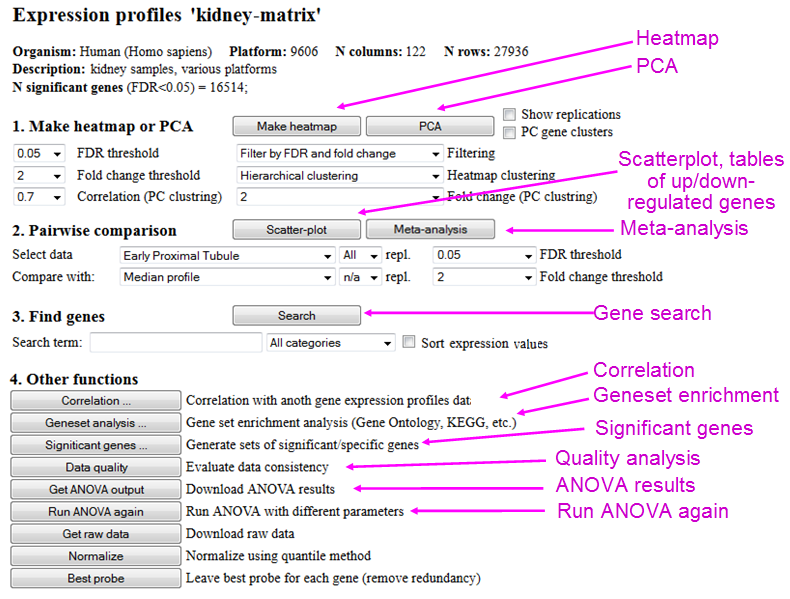
From this screen you can plot a heatmap, do Principal Component Analysis (PCA), make a scatterplot that displays differentially-expressed genes, and search for a specific gene to display its expression profile. Other functions (see sections 6 - 11) include meta-analysis, correlation with another gene expression profiles data, gene set enrichment analysis (e.g., for functional annotations), generating sets of significant/specific genes, evaluating data quality, downloading statistical results (ANOVA), downloading raw data, normalizing data with quantile method ( Bolstad et al., 2003), and removing redundant probes (leaving best probe for each gene).
Statistical analysis of gene expression data is based on the single-factor ANalysis Of VAriance (ANOVA). The program calculates F-statistics which is a ratio of factor variance (i.e., variance between averages for factor levels) to the error variance. F-statistics is then used to estimate the P-value according to theoretical F-distribution. Because in microarray analysis we simultaneously evaluate changes among several thousands of genes, it is necessary to adjust results for multiple hypotheses testing. The False Discovery Rate (FDR) shows the expected proportion of false positives among genes that are considered significant; it is estimated from p-values using method of Bejamini-Hochnberg. FDR ≤ 0.05 and fold change ≤ 2 are used as default criteria of statistical significance. The error model attempts to get a better estimate for the true error variance than the error variance estimated from data (we call it 'empirical error variance'). In ExAtlas, we use the maximum of empirical error variance and error variance averaged across 500 genes with similar average expression. This error model was proposed in the NIA Array Analysis software as a method to reduce the number of false positives.
Additional options for running ANOVA are available if you chose to "Run ANOVA again" which is a button in the section "Other functions" near the bottom of the web page (Fig. 4). This button is not available for public data sets, but you can make your own copy of public data (hint: use "Edit" button for "expression profiles" in section "File management", Fig. 2; select all samples and save them as your own data set), and then run ANOVA with custom parameters or with a custom annotation file for the array platform. When running ANOVA again you can select one of the following error models: 1 = Actual error variance for each probe, 2 = Average error variance for probes with similar expression level, 3 = Bayesian correction of error variance (Baldi & Long 2001), 4 = Maximum between actual and expected average error variances, 5 = Maximum between actual and Bayesian error variances. You can select a cutoff expression value (probes with maximum value below cutoff are ignored), modify threshold z-value used to remove outliers, modify proportion of probes with high error variances to ignore in error models, or modify the number of probes in a sliding window to average error variance. In addition you can set an option to use probe ID if gene symbol is missing. This option allows processing of non-annotated probes.
If the input file has no replications, then the error variance is estimated based on the assumption that at least half of gene expression values (log-transformed) represent random deviations from the average, and less than half values correspond to the effects of factors. First, genes are sorted according to their average log-expression, and then error variance is estimated within a sliding window of 500 genes with similar expression. Absolute deviations of log-expression of each gene from its mean expression value in all samples (i.e., |x-M|) is then combined for all 500 genes into one data set. For example, if the data matrix has 15 columns (=samples), then there will be 7500 deviation values (15 x 500) within the sliding window. The error variance is then estimated as the median of these deviation values divided by 0.675 (which is inverse half-normal cumulative distribution for 0.5):
In ExAtlas, ANOVA is run when you open the gene expression profile matrix for the first time. If the file has too many samples, the interruption screen (Fig. 3) may appear to wait until the task is finished. A tab-delimited text file with ANOVA results can be downloaded by clicking the button "Get ANOVA output" at the bottom of the screen which appears after you open any gene expression matrix file (see Fig. 4).
3.3. Plot a heatmap for the gene expression profile matrix
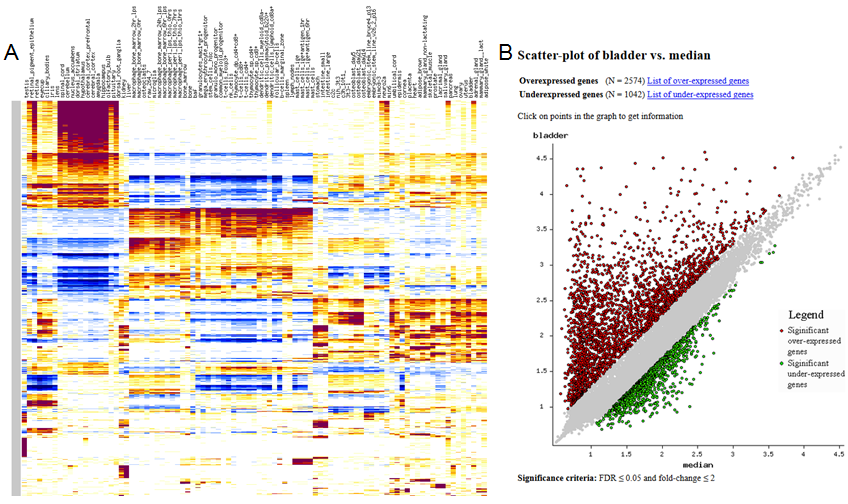
To plot a heatmap (see example in Fig. 5A), select gene filtering parameters (FDR threshold
and fold change threshold), kind of filtering, and kind of clustering in the upper portion of
the "Open expression profile" screen (Fig. 4). You can check the box "Show replications" if you
want to see data for individual replications. Then click the button "Make heatmap".
Filtering of genes is important, first, to save processing time, and second, to make the heatmap
easier to view. Non-significant genes (FDR<0.05) only add noise to the heatmap, and better
filtered out. Fold-change threshold = 2 is recommended. After the heatmap is displayed,
you can download the filtered and sorted matrix (as a tab-delimited text file) by using the link
"Matrix file" at the top of the page. This file can then be examined in Excel. Because of the
large number of genes, gene symbols may be not visible and represent by gray area (or lines) at the
left side of the heatmap. However, if you click in the row header area, gene name and expression profile
will be displayed.
The bottom portion of the screen is designed for editing the heatmap. For example, you can change the maximum value and click "Re-plot the matrix" button. If the maximum value is reduced, the colors will become darker, if the maximum value is increased, the colors will become lighter. Then, you can delete of move columns and rows using menu fields. For example, you can select a column (or a column range) and move it before another selected column.
3.4. Principal Component Analysis (PCA)
Principal component analysis (PCA) can be launched using the same filtering
parameters as for heatmap generation. You can check the box "Show replications" if you
want to see data for individual replications. Click the button "PCA" located near the top of the
"Open expression profile" screen (Fig. 4). PCA is computed using the Singular Value Decomposition
(SVD) method that generates eigenvectors both for rows and columns of the log-transformed
data matrix (Gabriel 1971. Biometrika 58: 453-467; Chapman et al. 2002. Bioinformatics. 18: 202-204).
For plotting of tissues and genes (biplot) we used column projections. The advantage of the
biplot compared to a traditional PCA is that the user can visually explore associations between
genes and tissues. ExAtlas generates 2-dimensional and 3-dimensional
(based on VRML) biplots (Fig. 6). All biplots (including 3D) are interactive; each gene is
a hyperlink to its annotation and expression pattern. To view PCA in 3-dimensions you need a VRML
viewer, for example FreeWRL or Cortona3d.
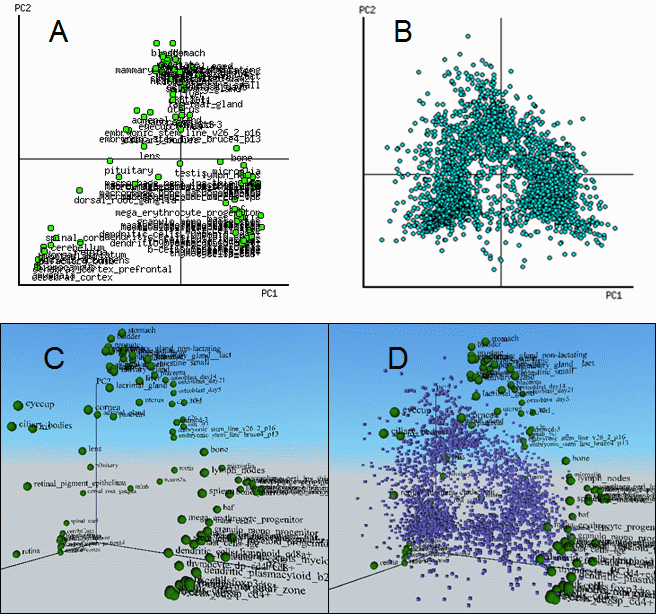
Fig. 6. PCA and biplot of mouse gene expression in various tissues (GNF database). A and B = 2-D biplot for tissues and genes, respectively; C = 3-D PCA; D = 3-D biplot for tissues (green spheres) and genes (blue cubes).
Checkbox "PC gene clusters" (Fig. 4) is used to identify 2 clusters of genes that are positively and negatively correlated with each principal component (Fig. 7). The degree of gene expression change within a specific PC is measured by the slope of regression of log-transformed gene expression versus the corresponding eigenvector multiplied by the range of values within the eigenvector. Gene is associated with the most correlated PC; however two additional conditions should be met: (a) the degree of gene expression change exceeds the threshold (default = 2-fold change), and (b) the absolute value of correlation exceeds the threshold (default = 0.7). These two parameters can be changed in the menu: "Correlation (PCA cluster)" and "Fold change (PCA cluster)" (Fig. 4).
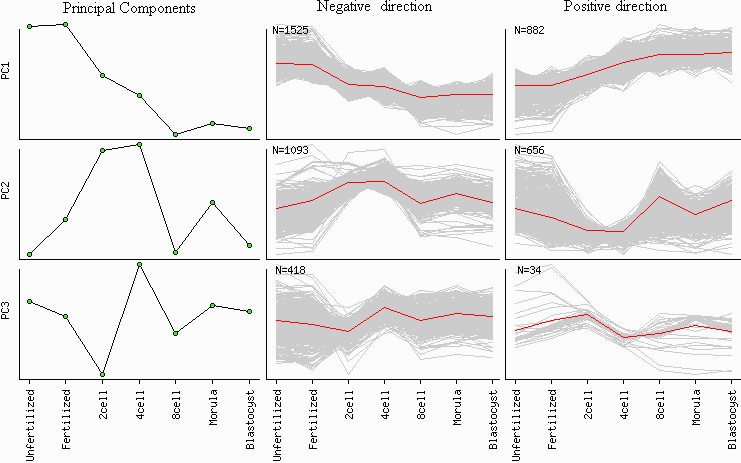
Fig. 7. Gene clustering based on principal components
3.5. Search for a gene and display the expression profile
Type a gene symbol or GenBank accession number in section "Find genes" near the middle of the
"Open expression profile" screen (Fig. 4) and click the button "Search". You can specify
what category of gene description you search (gene symbol, GenBank accession, gene name, probe ID).
If many genes (or probes) match to your search, all of them
will be displayed, and then you can select individual genes or probes. Checkbox "Sort" can be checked
if you wish to sort cell/tissue types according to the expression of the gene you search. When the gene
(or probe) is found, ExAtlas generates a histogram with gene expression profile (Fig. 8),
and a table of expression in each tissue/cell type. The histogram shows average log-expression values
for each cell type or tissue relative to the global median; to see values for individual replications
click the button "Show replications".
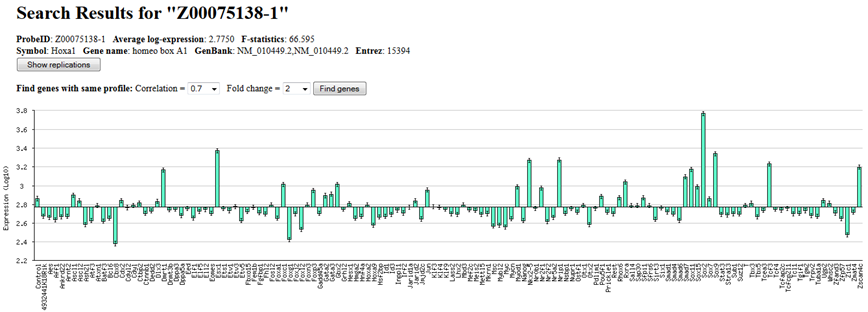
Fig. 8. Expression change of Hoxa1 after induction of 137 transcription factors in mouse ES cells
From the screen with gene expression histogram (Fig. 8) you can search for other genes with the same expression profile using correlation threshold and fold change threshold (which are applied simultaneously).
3.6. Pair-wise comparison of expression profiles of tissues or cell types
Select two tissues or cell types which you want to compare from pull-down menu in the section
"Pairwise comparison" near the middle of the "Open expression profile" screen (Fig. 4). As a baseline,
you can use median expression instead of a second tissue. Then select parameters (FDR threshold
and fold change threshold) and click the button "Scatter-plot". The scatterplot is a graph where
each point represents one gene with x-coordinate = log expression in tissue #2 (or median) and
y-coordinate = log expression in tissue #1. Gray dots = non-significant genes, red dots = significant
upregulated genes, and green dots = significant downregulated genes. Statistical significance is
based on z-value which are estimated from the ANOVA error variance by equation:
| z = |m1 - m2| / sqrt[ErrVar*((1/n1)+(1/n2))], |
If you use median expression profile for comparison (as control) then an additional feature is recorded in the output table: a z-value that characterizes gene specificity (column header "Specificity"). This z-value is estimated by comparing log-expression in a given tissue (mi) with average expression in other tissues (M) that are not correlated with this tissue (see details here).
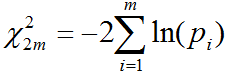
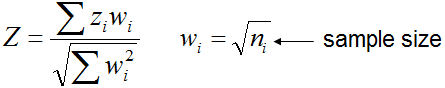
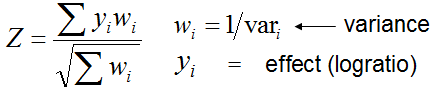
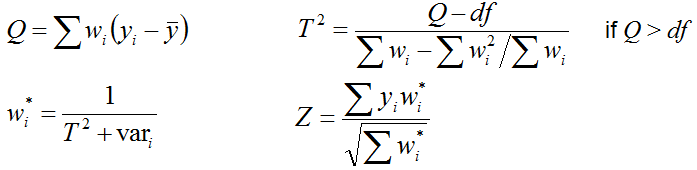
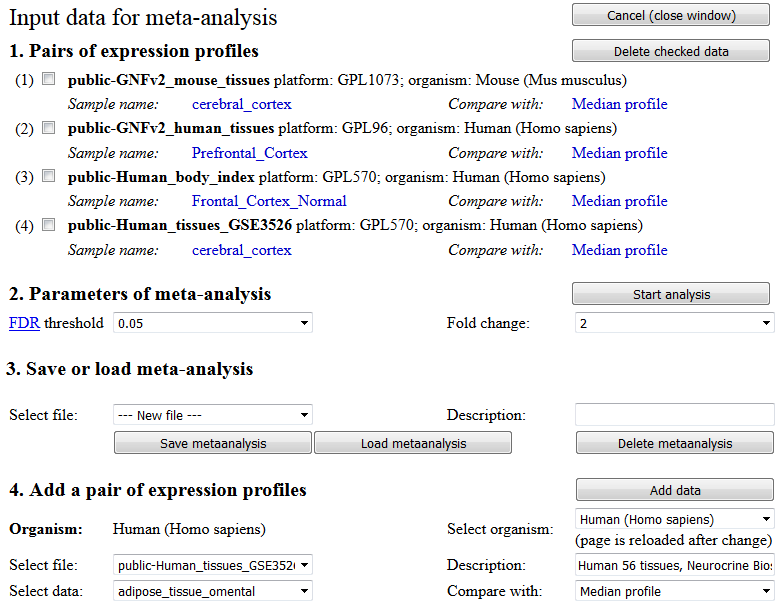
When all data sets for meta-analysis are assembled, select parameters for meta-analysis
(FDR threshold and fold-change threshold), and click the button "Start analysis". The
output page shows the number of significant genes for each method of meta-analysis. Click
on the number of gene to display the list of genes and corresponding statistics. Effects are
shown as either logratio (log10) (default) or as fold change. The format of effects can be
selected above the output table that shows the number of significant genes for each method.
The list of significant genes can be further explored for significant overlap with various
data sets.
3.8. Correlation between different gene expression data sets
To characterize the effect of treatments on gene expression profiles it is often necessary to
examine correlations between different gene expression data sets. For example, the change of
expression of genes following the induction of various individual transcription factors in ES
cells was compared with gene expression profiles in various tissues and cell types
Nishiyama et al. 2011. Results indicated
that some transcription factors (e.g., Ascl1, Gata3, Myod1, Sfpi1) induced tissue-specific genes.
To estimate correlations, open the first file with gene expression profiles, then click the
"Correlation" button in the section "Other tasks" (Fig. 4). This will take you to the next
screen where you can select the second file with gene expression profiles (Fig. 10). The second file can
be the same as the first one if you wish to generate an auto-correlation matrix. If you want
to compare gene expression change between different species, then select a species
for comparison. The screen will be reloaded with a list of data for that species. Use FDR threshold
and fold change threshold to limit the number of genes. Lower values of FDR and higher values of
fold change correspond to more stringent filtering.
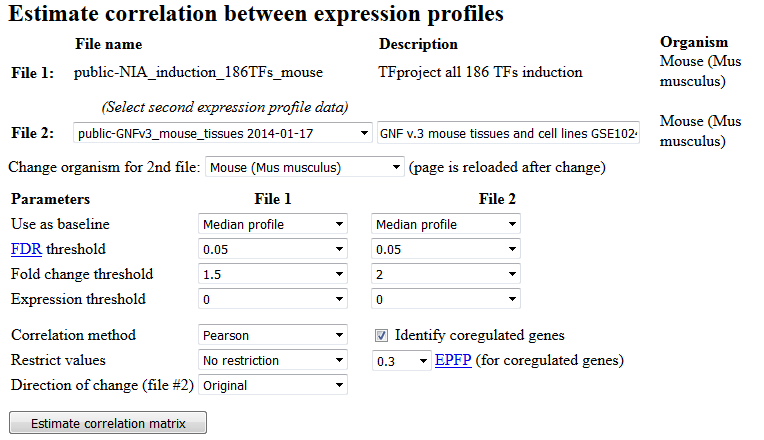
Fig. 10. Screen for correlation analysis of two data sets with gene expression profiles.
The algorithm for estimating correlations is the following.
If you check the box "Identify coregulated genes", then ExAtlas will identify lists of genes that are both upregulated or both downregulated in two data files if correlation is positive and significant (z ≥ 2), and Expected Proportion of False Positives (EPFP) is smaller than specified threshold (default threshold = 0.5). The algorithm for finding positively coregulated genes is based on the analysis of data points in the positive quadrant (i.e. x>0 and y>0). Negatively coregulated genes are identified in the same way in the negative quadrant. First, logratios of gene expression change are all replaced by their rank. If null-hypothesis is true (no correlation) then the genes are expected to have a uniform random distribution in the positive quadrant (Fig. 11A). To estimate EPFP for a gene with rank rx in the first expression profile (file #1) and rank ry in the first expression profile (file #2), we estimate the density of dots/genes in a rectangle with lower left corner at (rx,ry) coordinates (Fig. 11B, dark-shaded area) and compare it with the density of dots in two adjacent rectangles to the left and down (light-shaded areas). EPFP equals the density of dots in the light-shaded divided (which serves as a baseline) by the density of dots in the dark-shaded area. Because we have two light-shaded rectangles, EPFP is estimated twice, and then we select the larger value (to be conservative in our assessment). Because EPFP may not monotonically decrease with increasing rank rx and ry, it is forced to decrease monotonically. In particular, if EPFP(rx1,ry1) > EPFP(rx,ry), and rx1 > rx, and ry1 > ry, then EPFP(rx1,ry1) is set equal to EPFP(rx,ry).
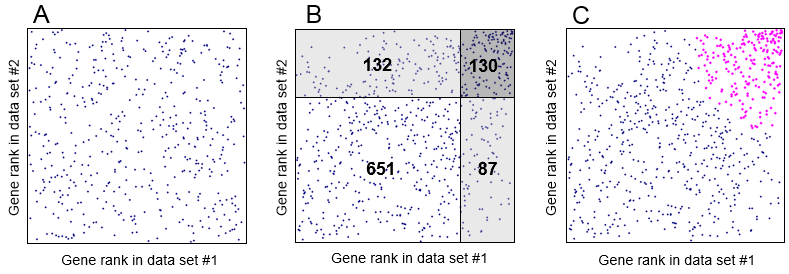
Fig. 11. Estimating Expected Proportion of False Positives (EPFP) for coregulated genes: (A) scatter-plot of gene expression rank in the positive quadrant if there is no correlation. (B) The same plot if gene expression profiles are correlated, numbers indicate gene counts. The density of dots/genes in the dark-shaded rectangle, 130/(132+130)/(130+87)=0.002287, is compared with the density of dots/genes in two light-shaded rectangles: 132/(132+130)/(132+651)=0.000643 and 87/(651+87)/(130+87)=0.000543. Two estimates of EPFP are generated for the gene at the low left corner of the dark shaded rectangle (with expression ranks rx=651+132=783 and ry=651+87=738): EPFP1 = 0.000643/0.002287 = 0.281 and EPFP2 = 0.000543/0.002287 = 0.237. The greater value is selected: EPFP = 0.281. (C) All coregulated genes with EPFP≤0.3 are highlighted (magenta).
To identify oppositely coregulated genes (i.e. upregulation in file #1 associated with downregulation in file #2 and vice versa), set "Direction of change (file #2)" to "Reversed" (Fig. 10). Then gene expression change for File #2 is inverted (multiplied by -1).
3.9. Exploring the output file
When the output table for correlation analysis is generated, results are saved in the output file,
which is opened automatically. Output files can also be opened manually from the main menu
from the pull-down selection list (Fig. 2); after selecting file click the "Open" button. When
output screen is displayed (Fig. 12), then it can be used to
plot the full output table as a heatmap (section #1) or to plot bar charts for rows and columns
of the output table (section #2). Examples of output graphs are shown in Fig. 13. When plotting the full table, select which values to plot.
Plotting options depend on the type of analysis and generally include z-values, which indicate
the significance of correlation. In addition, correlation values and/or the number of
associated genes is provided. After you selected which table to plot, click the button
"Plot output table". You can also plot profiles for individual rows and columns of the
output table by selecting respective rows or columns in section #2 "Profiles of rows,
columns, and cells". Values are sorted in profiles from high to low because sorting is convenient
for functional annotations of genes (e.g., Gene Ontology or pathways).
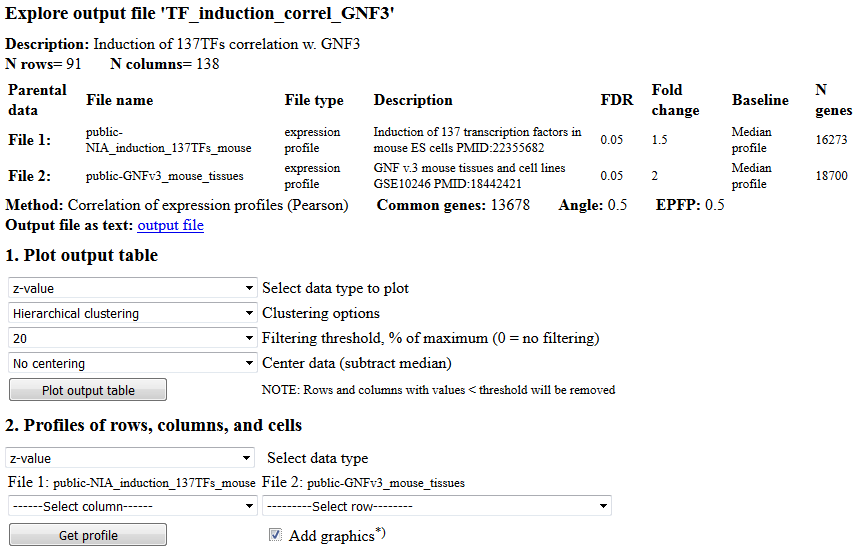
Fig. 12. Open output file screen: results of correlation analysis.
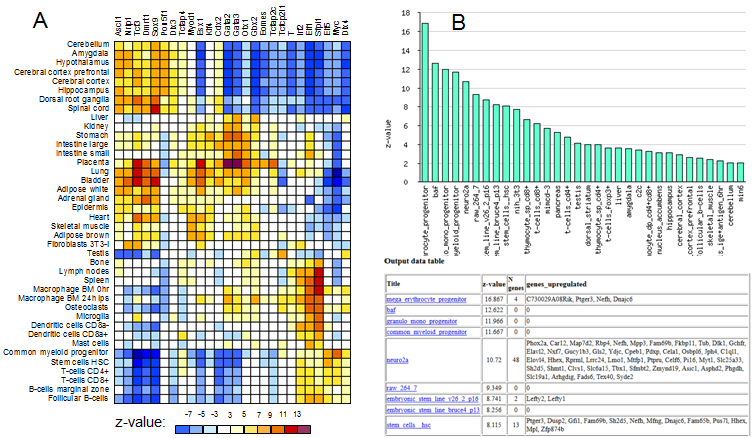
Fig. 13. Example of correlation matrix (A) and profile for a single row/column (B).
3.10. Geneset enrichment analysis of up/down-regulated genes
Geneset enrichment analysis is used to evaluate if specific genesets (such as Gene Ontology
or KEGG pathways) are over-represented among upregulated and/or downregulated genes. The
advantage of geneset enrichment analysis compared to a simple overlap of
genesets is that no thresholds are used for selecting differentially expressed genes.
In particular, geneset enrichment analysis can find significant associations with functional
genesets even if there are no significantly upregulated genes based on standard criteria
(e.g., FRD $le; 0.05 and change ≥ 2 fold). Among various existing methods for geneset
enrichment analysis we use Parametric Analysis of Gene Enrichment (PAGE) (Kim & Volsky 2005, PMID:15941488)
because of its simplicity and reliability (Zhang et al. 2010, PMID: 20092628). PAGE is based
on the comparison of the average expression change in a specific subset of genes,
xset, with the average expression change in all genes, xall:
| z = (xset - xall)*sqrt(nset)/SDall, |
Fig. 14. Screen for starting geneset enrichment analysis (PAGE)
To start PAGE analysis, select the geneset file using pull-down list (Fig. 14). To use geneset file for a different species, first select species. The screen will be reloaded with a list of data for that species; after that select the geneset file. To identify associated genes (e.g., target genes with binding sites of transcription factor which at the same time responded to the induction or knockdown of the same transcription factor) check the box "Identify associated genes". Use EPFP threshold and fold change threshold to limit the number of associated genes. Lower values of EPFP and higher values of fold change correspond to more stringent filtering.
Viewing the output file is similar to that for correlation analysis. You can plot a matrix heatmap or profile for individual columns or rows. If associated genes were identified they will appear in the profile (as in Fig. 13B). If the list of genes is too long it is truncated. To see the full list of genes (Fig. 15A), click on the row header. In addition, at the end of the list you will find a rankplot that shows graphically the enrichments of genes that belong to the given geneset among either upregulated or downregulated genes (Fig. 15B).
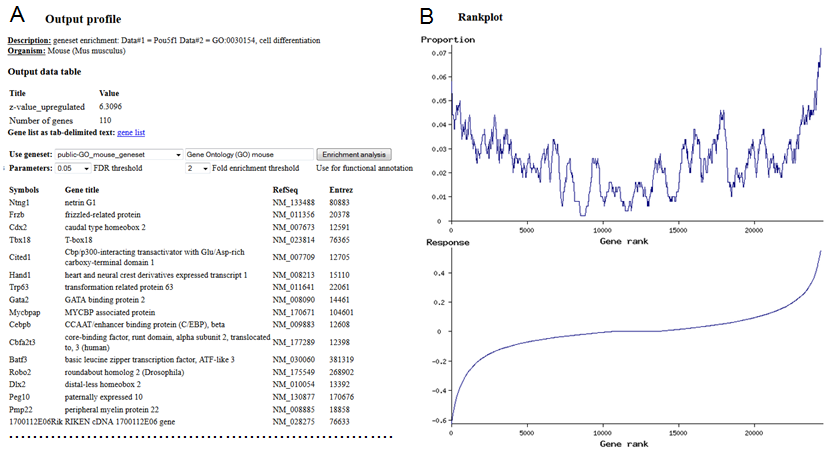
Fig. 15. List of associated genes (A) and a rankplot (B). In this specific case, genes from geneset are enriched among both upregulated and downregulated genes, but more strongly - for upregulated genes.
3.11. Generate a file with differentially-expressed genesets
ExAtlas automates the generation of genesets of upregulated and downregulated genes, which can
be later used for comparison with other data sets. Expression of each gene is compared to the
baseline expression, which can be selected as a median expression value (default) or expression
in some specific tissue/organ or cell line. Conditions of
statistical significance are defined by FDR threshold and fold change threshold. Additional
condition is gene specificity which allows to narrow down the list of genes to specific genes
only. Specificity is measured by z-value, as explained in the pair-wise comparison
section. To select highly-specific genes use z-values ≥ 6. Before starting the task, don't forget
to edit the name and description of the output geneset file, then click the button "Save
significant genes". When the task is finished, the output file displays a histogram of the number
of significantly upregulated (orange) and downregulated (dark blue) genes (Fig. 16).
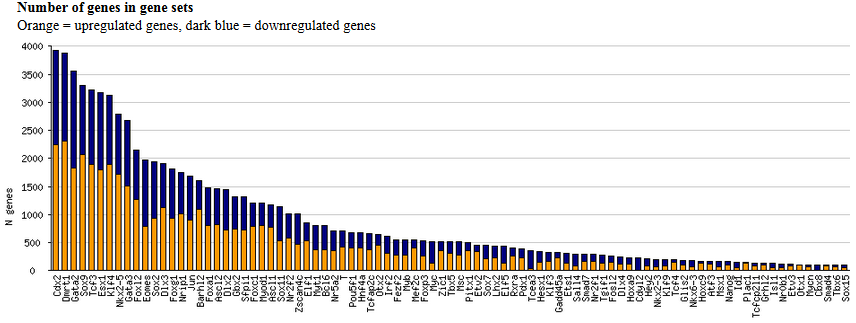
Fig. 16. Histogram of the number of significantly upregulated (orange) and downregulated (dark blue) genes after the induction of various transcription factors in mouse ES cells.
3.12. Explore a geneset file and/or analyze gene overlaps with another file
When you open a geneset file from the main menu (Fig. 2), a new window appears which allows the
user to find a geneset with a specific name/description (use button "Search") or select a
geneset from the alphabetically ordered list of all genesets (use button "Display genes") (Fig. 17).
The second portion of the menu is designed for starting the analysis of geneset
overlap. You can either select another geneset file and
simply paste a list of genes into the provided text area. Then select parameters
of statistical significance (FDR threshold and fold enrichment threshold) and click the button
"Overlap analysis". The program identifies common genes for each pair of genesets from
the first and second geneset files. And if the number of overlapping genes is greater than
expected by random, then it uses hypergeometric distribution to evaluate the significance of
gene enrichment. This is the traditional way of analyzing gene enrichment which is a simpler
alternative to a more sophisticated PAGE method described above.
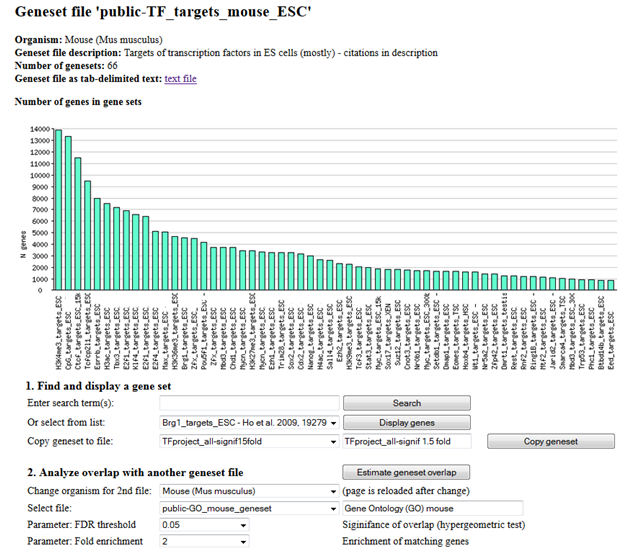
Fig. 17. Open geneset of targets of transcription factors in mouse ES cells.
When a specific geneset is selected, then the full list of member genes is displayed. From this screen you can test the significance of overlap with any other available geneset data, such as GO, KEGG, etc.
3.13. Evaluate quality of samples and remove low-quality samples
Click the button "Data quality" near the bottom of the "Open expression profile" screen (Fig. 4) to
run quality control program. If the data file is large, the interruption screen (Fig. 3) may
appear as discussed above. Quality control checks (a) correlation of log10-transformed expression
of housekeeping genes with standard data (RNA-seq), and (b) consistency between
replications. Consistency of replications is assessed by modified standard deviation (SD) of
the log-transformed expression in each sample from the tissue-specific median (where outliers with
z > 3.5 are not used for estimating median). In general, SD < 0.1 means good quality, and
SD > 0.3 means bad quality. Correlation of expression of housekeeping genes usually is in the
range from 0.5 to 0.95. If it falls below 0.5, then the quality may be low. Checkboxes
located near each sample allow the user to select samples with low quality for deletion.
3.14. Upload files for analysis (formats, normalization, etc.)
The "Upload data file" button in the main menu (Fig. 2) is used to open the screen for
file upload (Fig. 18). You either browse for the file to be uploaded (button "Browse..") or paste the
text file into the provided text area. Then, select the type of file (i.e., Gene expression
profile matrix, Gene set file, Samples file, List of geneset, Output file, or Annotation file).
If you want to store the file under different name, type-in the file name in the "Rename file as:"
field. Fill-out file description. If the file with gene expression profile table does not
include information on array platform, then you need to select array platform.
If the array platform is not present in the pull-down menu list, you need to upload a file with
platform annotation which should include at least 3 columns: "probe ID", "gene symbol", and "gene name".
You can add more columns that specify GenBank accession numbers, Entrez ID, or Unigene ID.
If gene symbols or GenBank accession numbers are used as probe ID, then select "Gene symbols" or
"public-genebank" platform annotation, respectively.
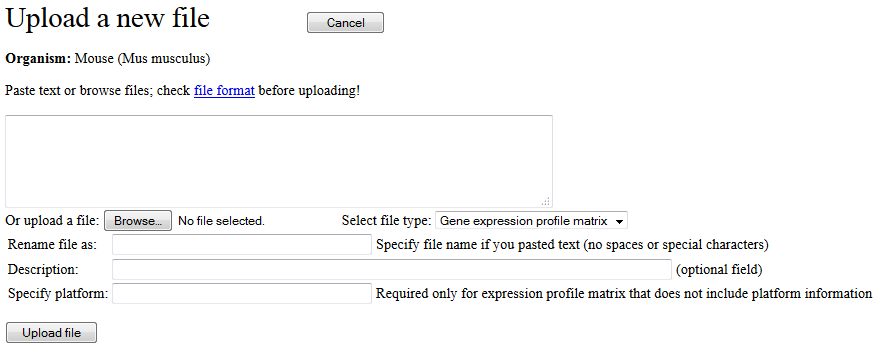
Fig. 18. Screen for uploading custom data files.
Here is a brief description of file formats.
The gene expression profile is a tab-delimited text that follows MIAME standards. All matrix
files downloaded from GEO can be directly uploaded to ExAtlas. The file has header lines that
start with "!" sign. However, these lines are optional. You can upload a file even without these
lines if you specify platform for the gene expression profile file.
Header lines are followed by a table with data lines that specify the intensity of feature
signals. Here is an example of a gene expression profile matrix file:
!Series_title "Gene expression of human soft tissue sarcoma" !Series_geo_accession "GSE2719" !Series_pubmed_id "15994966" !Series_summary "Gene expression profiles of 39 human sarcoma samples (GSM 52571-GSM52609)..." !Series_type "Expression profiling by array" !Series_platform_id "GPL96" !Series_platform_taxid "9606" !Series_sample_taxid "9606" !Sample_title "brain" "stomach" "colon" "pancreas" "prostate" ... !Sample_geo_accession "GSM52556" "GSM52557" "GSM52558" "GSM52559" "GSM52560" ... !Sample_taxid_ch1 "9606" "9606" "9606" "9606" "9606" ... !Sample_data_row_count "22283" "22283" "22283" "22283" "22283" ... !series_matrix_table_begin "ID_REF" "GSM52556" "GSM52557" "GSM52558" "GSM52559" "GSM52560" ... "1007_s_at" 2867.1 1780.8 1921.8 2486.1 4151.4 ... "1053_at" 216.4 196.8 145.3 127.1 109.7 ... "117_at" 135 121 157.2 162.6 267.8 ... "121_at" 916.1 1075.7 922 2192.9 1198.8 ... "1255_g_at" 149.8 35.5 32.7 96.3 47.6 ... .................................................................. !series_matrix_table_end
Sample names are taken from the line "!Sample_title" or from the line of column headers that follows after "!series_matrix_table_begin". Column headers for replication samples should be exactly matching (case-sensitive). It is not required to reorder columns so that all replications are placed together; replicetion samples are recognized by column headres even if they are separated by other samples in the table. ExAtlas can process 2-dye arrays that use reference RNA consistently as one of the channels (e.g., Cy5 or Cy3). In this case, two columns that correspond to the same array (channel #1 and channel #2) should be placed together and the column representing reference RNA should be named "reference". If data are log-transformed or Z-value transformed, then select transformation type from the pull-down menu.
Because background subtractions may result in negative values, some array scanning programs avoid negatives by adding some constant value to signal intensity (e.g., 50 or 100). Usually this does not cause problems, but low-expressed genes may show weaker expression fold-change. If you would like to remove this constant value, then select "adjustment" value from the pull-down menu.
After you upload a new gene expression profile it will appear in the main menu. When you try to open it for the first time, it will run ANOVA (which may take some time).
Alternatively you can compile gene expression data column-by-column from one or multiple tab-delimited text tables. To use this option, select "Compile expression profile" option from the pull-down list "Select file type:". Type-in file name in the field "Rename file as" and description. Select array platform if applicable, then browse to select the first data table and click "Upload" button. After the table is parsed and column headers displayed on the screen, select columns to be extracted, specify their usage (Probe ID/tracking ID, Gene ID/name, or Gene expression), and possibly edit column header. If you have specified array platform, use column with probe ID as "Probe/tracking ID". Alternatively, select a column as Gene ID/name if it has gene symbols, GenBank acc., Entrez gene ID, or Ensembl gene ID. Please, edit column headers as 'symbol', 'refseq', 'genbank', 'entrez', or 'ensembl'. Probe/tracking ID or Gene ID/name should be common for all data files that are assembled together. When these data are uploaded, you can choose another data table and extract data from it until all data are compiled. It is necessary to specify Gene ID/name at least in one of the tables. For example you can upload an annotation table where both Probe ID/tracking ID and Gene ID/name are present. At any time you can edit sample names to make them meaningful and ensure that replications have exactly the same sample names (case-sensitive). If you have 2-dye arrays and one channel is used for reference RNA, then edit column name as 'reference'. In this case reference expression will be used for normalization as follows: norm(x) = x*My/y, where x is signal intensity for sample, y is signal intensity for reference, and My is geometric mean of all reference values.
In a geneset data file (tab-delimited text), each line corresponds to one geneset. First item is geneset ID, the second is geneset description (which may be blank or duplicate ID), followed by all genes that belong to this geneset. Because some lines are rather long, geneset files may not always be opened in Excel. Geneset file may include header lines that all start with "!". Here is example of a geneset file:
CITRATE_CYCLE_TCA_CYCLE CITRATE_CYCLE_TCA_CYCLE Idh3g Pdha2 Fh1 Suclg1 Idh2 Pcx Pdha1 Idh3b Sucla2 Mdh1 Suclg2 ... ETHER_LIPID_METABOLISM ETHER_LIPID_METABOLISM Pla2g4e Pla2g7 Pla2g12a Pla2g4a Lpcat4 Agps Pafah2 Pla2g3 Pla2g2f Ppap2a ... ..........................................................................................................An alternative acceptable format of geneset files uses comma-separated lists of gene symbols:
CITRATE_CYCLE_TCA_CYCLE CITRATE_CYCLE_TCA_CYCLE Idh3g,Pdha2,Fh1,Suclg1,Idh2,Pcx,Pdha1,Idh3b,Sucla2,Mdh1,Suclg2,... ETHER_LIPID_METABOLISM ETHER_LIPID_METABOLISM Pla2g4e,Pla2g7,Pla2g12a,Pla2g4a,Lpcat4,Agps,Pafah2,Pla2g3,Pla2g2f,Ppap2a,... ..........................................................................................................Sample files (tab-delimited text) have 4 columns: (1) series ID from GEO, (2) Platform ID, (3) Sample ID, and (4) sample title/name. Samples with identical titles within the same data series are considered as replications. Check title spelling, spaces, and character case, because in the case of mismatch replications will not be recognized. Example:
GSE6290 GPL1261 GSM144590 renal corpuscle GSE6290 GPL1261 GSM144591 renal corpuscle GSE6290 GPL1261 GSM144594 Early Proximal Tubule GSE6290 GPL1261 GSM144595 Early Proximal Tubule GSE6290 GPL1261 GSM144596 Medullary Collecting Duct GSE6290 GPL1261 GSM144597 Medullary Collecting Duct GSE6290 GPL1261 GSM144603 sshaped_body GSE6290 GPL1261 GSM144604 sshaped_body GSE6290 GPL1261 GSM144605 sshaped_body ............................................................Annotation file has at least 3 columns: (1) Probe ID, (2) Gene symbol, and (3) Gene name. Additional columns may show accession number, Entrez, Ensembl, Unigene or other IDs. Do not use multiple gene symbols in the second coumn! If a probe matches to multiple symbols then select the best symbol for annotation. If you need to show other matching gene symbols, then make multiple copies of the line with this probe ID in the gene expression profile data and modify probe ID (enter unique new ID) which will be associated with alternative symbols. Annotation file always has a line with column headers and may include optional header lines that start with "!".
NIA-oligo Gene symbol Gene name GenBank Entrez Z00000225-1 Wdr74 WD repeat domain 74 NM_134139.1,NM_134139.1 107071 Z00000233-1 Tro trophinin NM_001002272.2,NM_001002272.2 56191 Z00000238-1 Edf1 endothelial differentiation-related factor 1 NM_021519.1,NM_021519.1 59022 Z00000241-1 Pfn1 profilin 1 NM_011072.2,NM_011072.2 18643 Z00000244-1 Rabep1 rabaptin, RAB GTPase binding effector protein 1 AK163126.1,AK163126.1 54189 .........................................................................Output files may include one or several tab-delimited tables. When you perform any analysis in ExAtlas (correlation, gene enrichment, significant genes, etc.) you can then download the output file to explore its format. Any tab-delimited table with first line of column headers and with the first column as row headers can be uploaded as output file for plotting as a heatmap. No additional formatting is needed.
Lists of genes (official gene symbols) can be uploaded to explore the enrichment of various genesets for functional annotations (e.g., for comparison with GO-terms, KEGG pathways). Genes can be formatted in one column or pasted as comma-separated text. After the list of genes is uploaded, select the geneset file for comparison (e.g., GO_mouse_geneset), specify parameters (FDR and fold enrichment) and click "Enrichment analysis". When the output opens, click on the button "Get profile".
3.15. Edit files
ExAtlas supports minor editing of uploaded files (except platform annotations). If you made
a mistake during file upload, you can fix it using the editing tool. In particular, users can
rename the file, edit its annotation, or specify a different microarray platform for gene
expression profiles. More editing options are available for gene expression profiles and
geneset files. In particular, users can select gene expression profiles (e.g., microarray samples)
or genesets and either delete them or copy to another file. If gene expression profiles are
copied to already existing file, then the user can select to co-normalize data in various
ways: (a) by quantile method, (b) by equalizing global median values for each gene, or (c) by
equalizing median values for selected samples within each data set. For example, if two projects
have data on gene expression profiles in normal liver, then the user can select all liver samples
in each data set and then use option (c). Options (b) and (c) represent batch-normalization
procedure which is often used for combining heterogeneous data sets. Because batch-normalization
generates better results than quantile method, we suggest not to combine different data series
from GEO in "Search GEO database" option, but to save each series
separately and later combine them using batch-normalization.
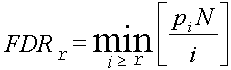
Feedback
Contact Alexei Sharov sharoval@mail.nih.gov
if you have problems with ExAtlas or suggestions for improvement.